3D-ICSS Analyses with ICSSgen3D and ICSScub3D
ICSSgen3D and ICSScub3D is end of support. The basic functionalities have been combined into py.Aroma, a multi-functional tool for aromaticity analyses. Please check the homepage of py.Aroma for more information.
Contents
- 1. Statement of need
- 2. Install and usage
- 3. 3D-ICSS analyses of 1-methylazulene
- 3.1 Create input files with ICSSgen3D
- 3.2 Submit input files to Gaussian calculation
- 3.3 Visualize 3D-ICSS with ICSScub3D
1. Statement of need
In the previous post, I introduced how to plot 2D-ICSS map to investigate the aromaticity. Sometimes, a 3D-ICSS is necessary if the molecular structure is complicate, and it may hard to be presented by 2D-ICSS map. Thus, I developed the 3D version of ICSSgen and ICSScsv, the Python programs kit: ICSSgen3D and ICSScub3D.
2. Install and usage
ICSSgen3D and ICSScub3D could be download from GitHub. To install and run the programs, please refer to the user manuals.
3. 3D-ICSS analyses of 1-methylazulene
3.1 Create input files with ICSSgen3D
3D-ICSS analyses of 1-methylazulene would be presented in this section. The calculation files in this section could be download from here. The optimization of 1-methylazulene is carried out at ωB97X-D/6-31G(d) level of theory with Gaussian 16 B.01 package.
An original input file is prepared, including the route line, calculation method, geometry coordinates. The original input file looks like:
%nprocshared=6
%mem=10GB
%chk=methylazulene.chk
#p nmr=giao rb3lyp/6-31g(d)
optimized_1-methylazulene
0 1
C -2.68122300 -0.69593600 0.00002600
...
Next, we need to determine the calculation region. In this calculation, the ghost would be added in the cuboid with X (-7.5, 7.5), Y (-6.5, 6.5) and Z (-6, 6).
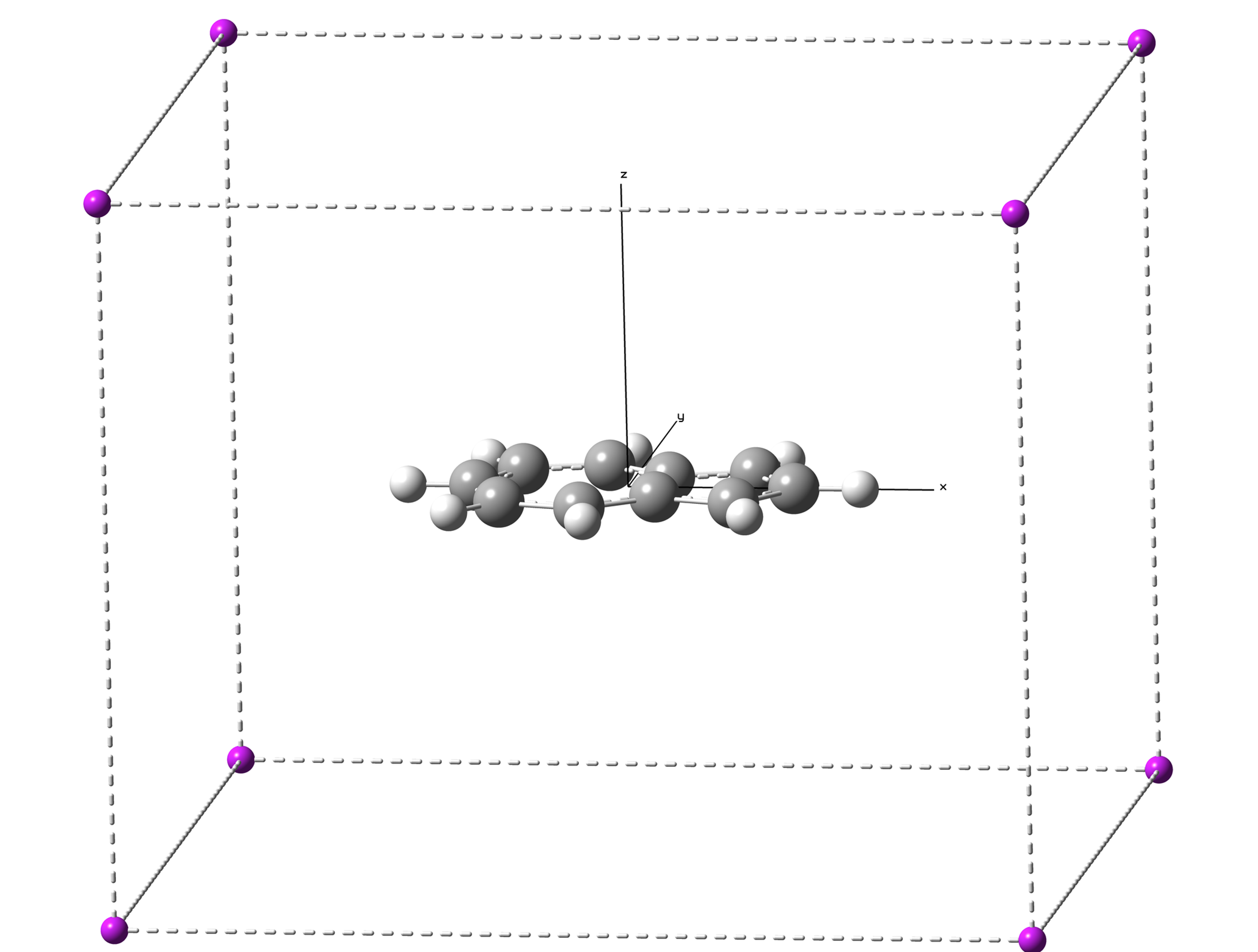
(For Gaussian 16, the maximum atom number is 8000, so, we need to separate the calculation into several input files. ICSSgen3D will add 7000 ghost atoms in each file, to make sure not to exceed the limitation.)
3.2 Submit input files to Gaussian calculation
ICSSgen3D would generate several input files, and we need to submit these input files to Gaussian calculation. The input files named from xxx_3DICSS_0001.log, xxx_3DICSS_0002.log, … . From the second input file, the guess=read keyword is included to save the calculation time by reading MO information from the previous .chk file. So, please save these input files into one folder.
I wrote a Bash Shell script (RunGJF) for submit all Gaussian jobs in the same dictionary one by one.
3.3 Visualize 3D-ICSS with ICSScub3D
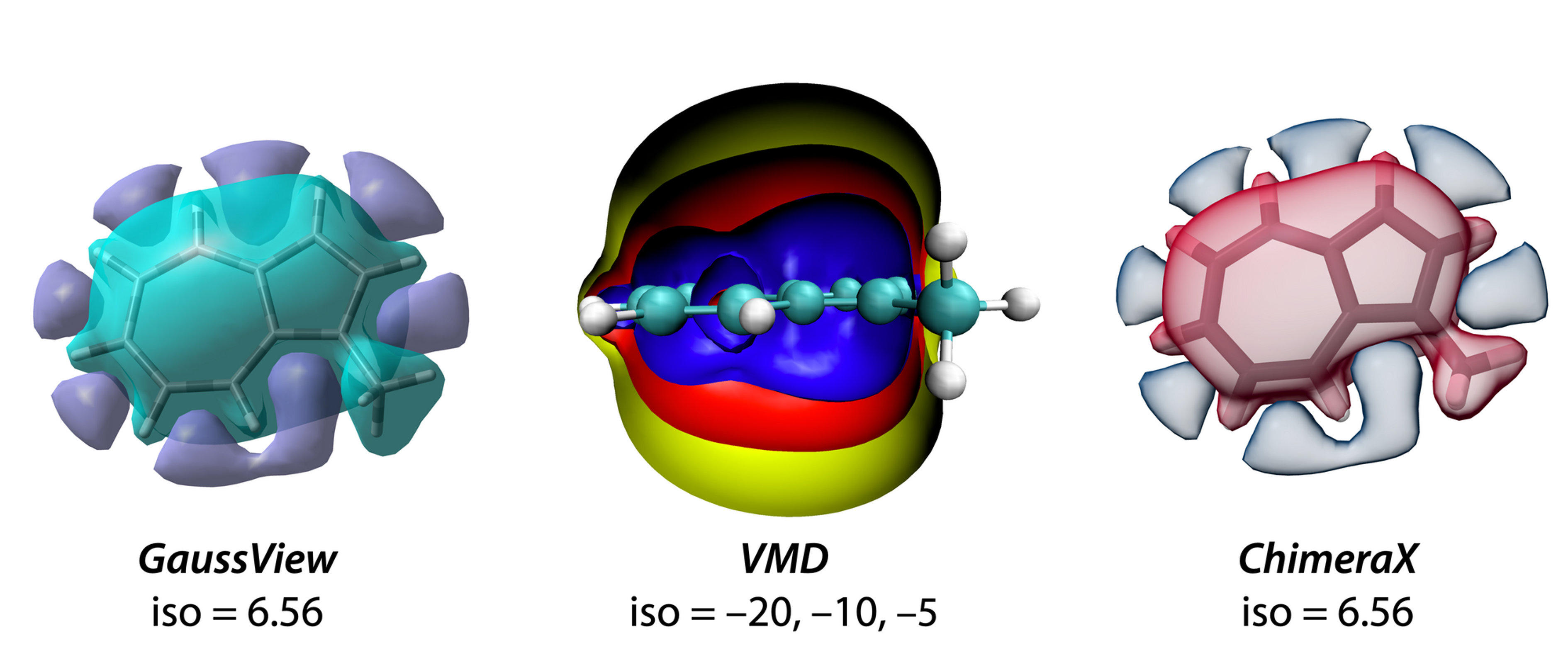
After finishing all calculation, we need to extract the magnetic shielding tensors from the output files. ICSScub3D can do this job for us. We just need to tell the output file path to ICSScub3D, and determine which component (isotropic, anisotropy, ZZ and so on) would be used for 3D-ICSS plot, the program can save the shielding tensor into .cub file automatically. We can visualize the 3D-ICSS plot with the .cub file using GaussView, VMD, Chimera X, etc.
Enjoy Reading This Article?
Here are some more articles you might like to read next: