2D-ICSS Analyses with ICSSgen and ICSScsv
ICSSgen and ICSScsv are end of support. The basic functionalities have been combined into py.Aroma, a multi-functional tool for aromaticity analyses. Please check the homepage of py.Aroma for more information.
Contents
1. Statement of need
2D isochemical shielding surface (2D-ICSS) maps, also known as 2D-NICS (nuclear independent chemical shift) maps, are useful tools for investigating the aromaticity of cyclic molecules. A large number of ghost atoms, in addition to the target molecules, must be included in the input file for 2D-ICSS calculations. After completing the calculations, the magnetic shielding tensors of all ghost atoms must be extracted from the output files. This process is a huge and tiresome task; therefore, we present ICSSgen and ICSScsv, two open-source, highly efficient, and user-friendly Python programs, to easily generate 2D-ICSS maps.
2. Install and usage
ICSSgen and ICSScsv could be download from GitHub. To install and run the programs, please refer to the user manuals.
3. 2D-ICSS analyses of 1-methylazulene
2D-ICSS analyses of 1-methylazulene would be discussed in this section. All files in this section could be download from here.
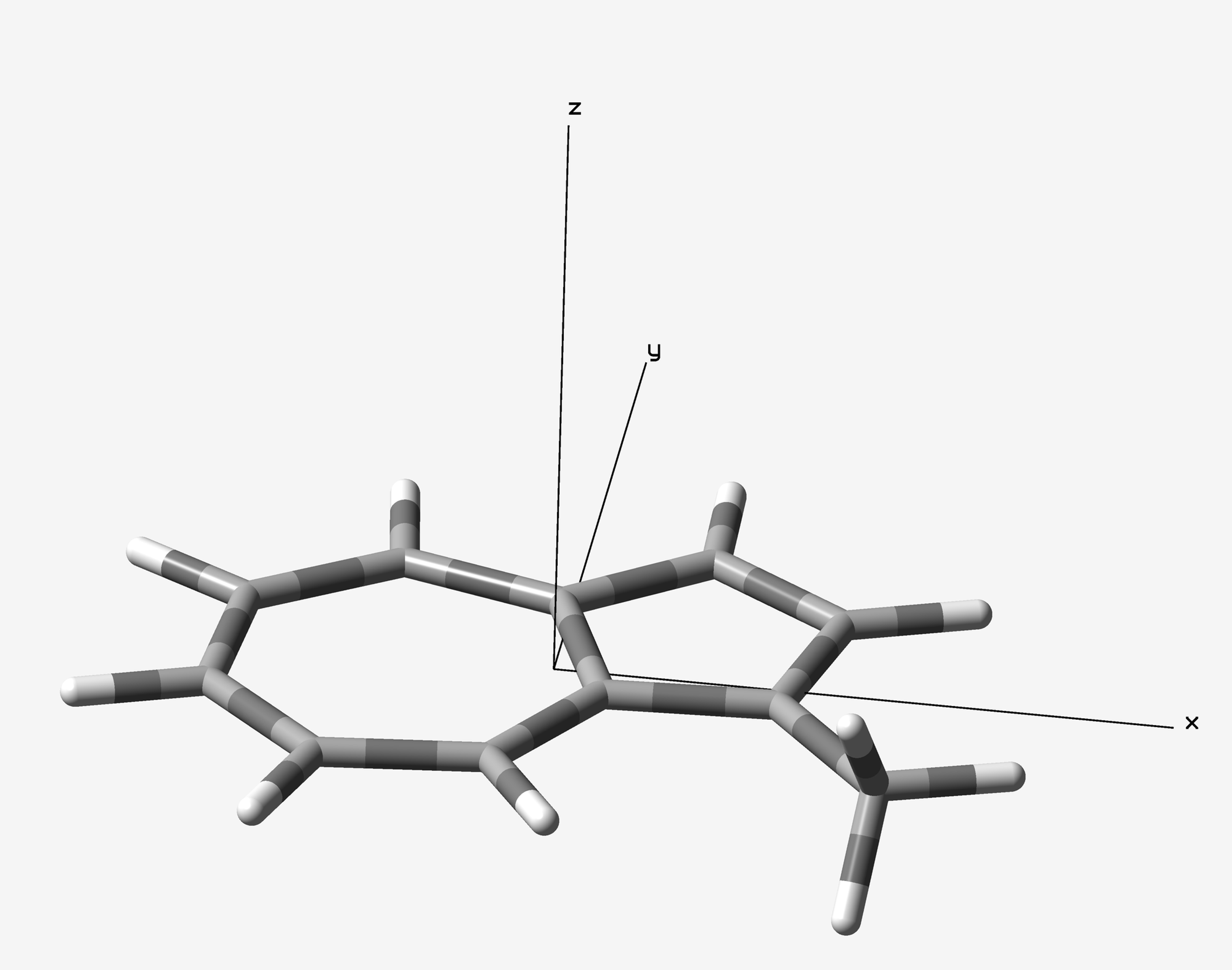
1-Methylazulene was optimized at ωB97X-D/6-31G(d) level of theory with Gaussian 16 B.01 package. The optimized azulene is located on XY plane (Figure 1). Two 2D-ICSS maps would be plotted on the XY plane with Z = 0 and Z = 1, in the range of X from -7 to 7 and Y from -6 6 Å. The input files were created by ICSSgen (Figure 2) and submitted to Gaussian calculations.

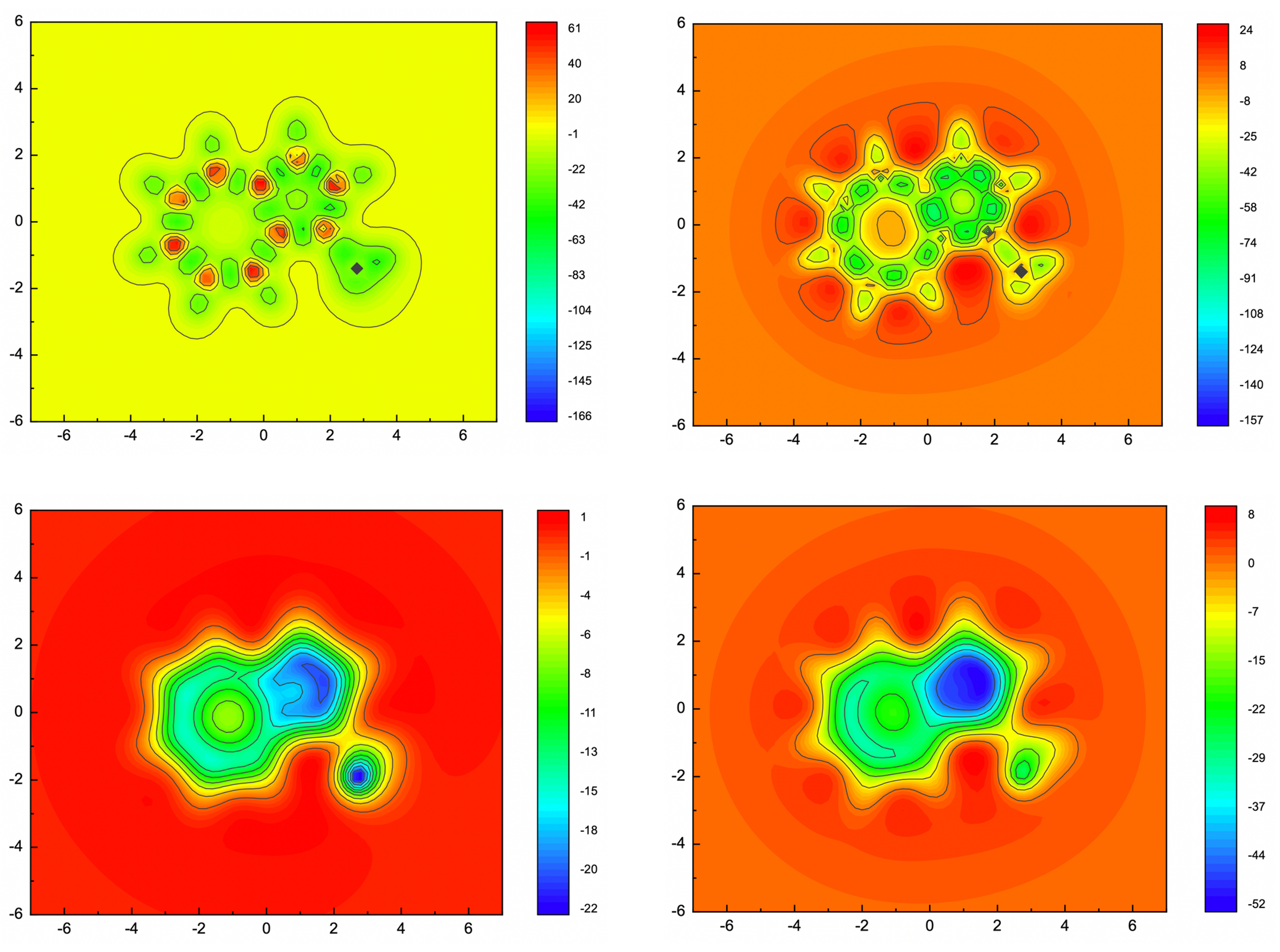
NMR (GIAO method) calculations were conducted at B3LYP/6-31+G(d) level of theory. The output files were then, processed with ICSScsv. The magnetic shielding tensors (isotropic and ZZ component) of ghost atoms were extracted to .csv files. 2D-ICSS maps were plotted with those date in .csv files by Origin (Figure 3). From these maps, the aromaticity of 1-methylazulene has been well proved by the negative shielding tensor values. Furthermore, the on-plane maps (Z = 1) show contribution from σ bonds, which sometimes annoying. The over-plane maps (Z = 1) well produced the contribution from π electrons.
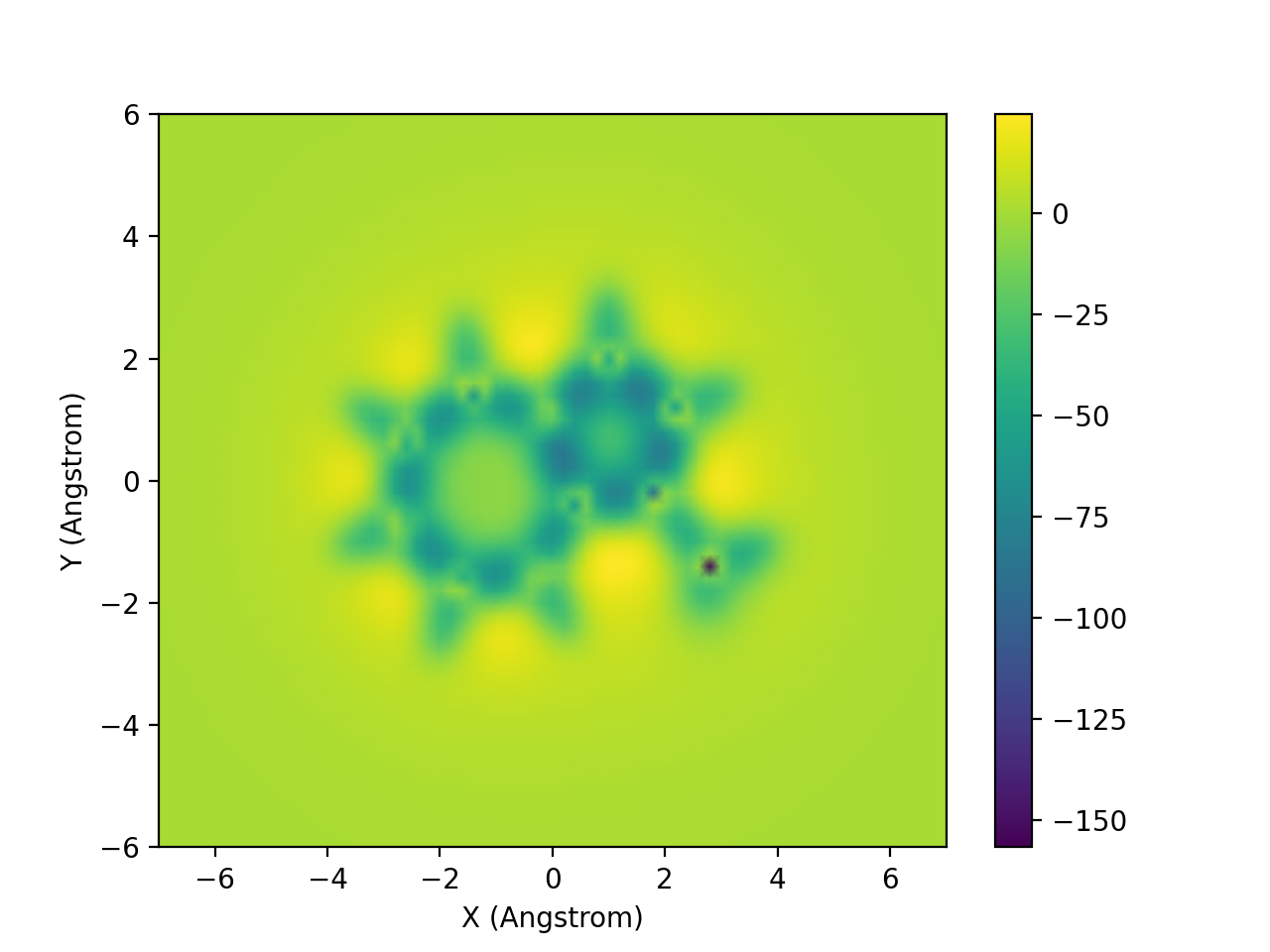
In the updated version 3.1, ICSScsv can save the 2D-ICSS image as .png file automatically after processing the output file. Python external library numpy and matplotlib were imported to plot the image (Figure 4).
Enjoy Reading This Article?
Here are some more articles you might like to read next: